A Fingerprint
of Our Genetic Makeup: DNA Typing

Deoxyribonucleic acid (DNA): A polymeric
molecule consisting of deoxyribonucleotide building blocks that in a
double-stranded, double helical form is the genetic  material
of most organisms. (courtesy
of iGenetics)
material
of most organisms. (courtesy
of iGenetics)
DNA is what sets
all organisms apart. The DNA that we
have is what makes us humans and the DNA horses have is what makes them
horses. Within humans as a species much
of the DNA molecule is identical but no two people have the exact same genome
and this is where DNA typing or fingerprinting comes into play. DNA typing techniques are used in criminal
trials, paternity tests, population genetics studies, proving pedigree status
for horses and dogs, and identification of dead bodies. Of course there are numerous other uses and
some will be explored later on.
The coding
regions of DNA (exons) are very similar between human individuals but there are
portions of DNA which seem to be noncoding regions which we know as introns. It is in these regions that repeated
sequences of base pairs exist which differ from one person to the next. These
sequences are called Variable Number of Tandem Repeats (VNTRs) if they are five
to a few tens of base pairs long. Short
Tandem Repeats (STRs) are two, three or four base pairs long and also show
great variation. Every individual has
some VNTRs (and STRs) and their VNTRs come from the genetic information donated
by their parents. The VNTRs can be
inherited from either that person’s mother or father and can be a combination
of both. A very important part of this
is that a person’s VNTRs will never have sequences that their parents do not
have. Scientists can use these regions
(VNTRs) to make a “fingerprint” to determine whether two DNA samples are from
the same person, related people, or non-related people.
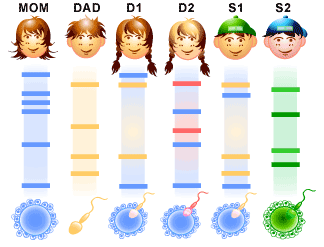
In the image to the left it is shown that Daughter one and Son one have VNTRs from both their mother and father. Daughter two has her mother’s VNTRs but not her father’s meaning she is from the mom’s other marriage. Son two has neither mom nor dad’s VNTRs because that son was adopted.
The two main processes for DNA analysis are:
1. Using restriction fragment length polymorphisms (RFLPs) detected by restriction enzyme digestion and Southern blotting analysis.
2. Using Length polymorphisms detected by PCR amplification and a dot blot assay.
 RFLP Identification
RFLP Identification
1. Isolate DNA from blood, semen, or any other cell sample.
2. Extract the DNA from the cell.
3. Digest the DNA into different sized fragments by restriction enzymes such as EcoR1.
4. Separate the DNA fragments by gel electrophoresis.
5. Denature the DNA strand into two single strands.
6. Transferring the DNA band pattern to a nylon membrane (also known as Southern Blotting).
7. Hybridization of the single strand of DNA with a radioactive probe which binds to specific sequences of DNA. (The final DNA fingerprint is built by using several probes simultaneously.)
8. Creating a picture of the radioactively marked DNA onto x-ray film.
After this is done you can visualize bands corresponding to the lengths of the fragments of DNA which can then be used to compare to another person’s for criminal trials or paternity tests.
PCR-based
Analysis
1. Isolate DNA from blood, semen, or any other cell sample.
2. Extract the DNA from the cell.
3. Amplify the DNA by using the polymerase chain reaction (PCR). This DNA is copied over and over again usually in a machine called a thermal cycler.
4. Treat the amplified DNA with a variety of probes that are bound to a blot (probes on a membrane are dipped into the amplified DNA). Each probe is found in a specific “dot” on the blot strip. A chemical reaction causes the dot to darken and become noticeable when DNA containing a particle variant is present.
5. Infer the genotype by the pattern of dots that indicates which probes the amplified DNA bound to.
A short preparation and analysis time of a few days makes this DNA typing technique more efficient then RFLP which take a few weeks.

There are
various other techniques for DNA typing.
The technique will be determined depending on the situation and the
amount of DNA available. All make use
though of the tandemly repeated sequences present in a person’s DNA. There are some issues though with the
accuracy of the tests and the possibility that the fingerprint may not be
unique to one individual. These
problems that arise and how they have affected the history of DNA
fingerprinting will be examined.
The History of DNA
Fingerprinting
I.
Tiselius,
1933
a. Invented electrophoresis for separating
proteins
II.
Frederick
Sanger, 1963
a. Developed sequencing procedure for
proteins
III.
P. H.
O’Farrell, 1975
a. Invented two-dimensional electrophoresis
IV.
E. M.
Southern, 1975
a. Published his procedure for testing the
existence of specific pieces of DNA
V.
Alfred J.
Jeffreys, 1984
a. Discovered the fundamental processes
behind DNA fingerprinting
b. Studied gene for myoglobin
c. Found non-functional “minisatelites” –
areas near gene which vary between individuals
d. When these segments were isolated,
enhanced, and radioactively labeled they could be used to distinguish
individuals through gel electrophoresis.
e. The original process could take up to six
weeks, but by 1991 Jeffreys had improved the test so that it took as few as two
days.
VI.
1985 –
First paternity test
VII.
1988 –
First criminal conviction based on DNA evidence
VIII.
1989 –
First conviction overturned based on DNA evidence
a. Gary Dotson served 10 years of his 25-50
year sentence
IX.
Kary
Mullis, 1993
a. Won the Nobel Prize for the development
of PCR (polymerase chain reaction) procedure.
b. Made isolation and analysis of DNA
quicker and easier by reducing the amount of DNA needed from the organism.
c. He originally conceived the idea in 1983.
X.
1996 –
First conviction based on mitochondrial DNA
XI.
1999 –
First “cold hit” obtained from a DNA database
a. Wallid Haggag is convicted of burglary
b. He was not a suspect, but police matched
blood found at the crime scene to Haggag’s blood which was on record in the
state DNA database
Problems with DNA
Fingerprinting
I.
Costs –
Time and Money
a. Can be very expensive and time-consuming
b. Less of a problem since Kary Mullis’s
development of PCR
II.
Chance
a. There are no clear matches – only
probabilities.
i.
The DNA
fingerprinting process examines several pieces of DNA, not the whole sequence
I.
Though
unlikely, two individuals could have the same DNA fragments while being
genetically distinct
II.
Racial and
ethnic groups are likely to have similar VTNR’s
III.
The Human
Factor
a. Comtamination
i.
DNA from a
lab technician or a police officer can become mixed with DNA from a suspect
b. The Jury
i.
DNA fingerprinting
can be confusing to explain to a jury.
ii.
Any doubt
about contamination can destroy the validity of DNA evidence
c. Misuse
i.
Planting
DNA evidence to frame a suspect
ii.
Concerns
about racial profiling stemming from similarities in VTNR’s
Applications of DNA fingerprinting/typing
1. Forensic
analysis:
DNA
typing can be used in forensic cases of murder, rape, homicide, and other
violent crimes. DNA samples are typically taken during a criminal
investigation, from the crime scene area or from the victims clothing or body
and include: blood, hair, skin cells, or any other genetic material. The
samples are then compared using VNTR patterns to determine a match either to
the suspect or victim. VNTR patterns can also be used to identify victims of
homicide when a sample from the parents is available. If the sample is
extremely small PCR can be used to amplify the DNA for the typing tests.
2. Population
genetics:
Used
to determine variability in various ethnic groups or populations.
3. Providing
pedigree status for certain animals:
Commonly
used to determine specific breeds of some horses and dogs. This DNA
fingerprint can be used for registering animals and establishing pedigree as
well as for parentage verification. DNA identity information can be used to correlate
EPDs to parent stock which allows selection of animals that meet set criteria
for performance.
Benefits
of Using DNA-based Animal Identity:
-Create a permanent
identity record for each animal
-Increase value based
on purebred or branded product verification
-Resolve pedigree
disputes
-Confirm Parentage in
multi-sire breeding programs
4. Forensic
analysis of wildlife crimes:
Can
be used to solve crimes against wildlife such as illegal hunting, trafficking
in endangered species, and the production and sale of products made from
illegally hunted animals. There is one lab in the world that deals with these
cases, The National Fish and Wildlife Forensics Laboratory in Ashland, Oregon.
They work on cases from the 50 United States and from 155 other countries
worldwide. The lab staffs 33 people that work on approximately 900 cases every
year. They have a reference warehouse that holds stuffed animals and animal
parts used to identify creatures. They have nearly 5,000 complete animals and
over 30,000 blood and tissue samples, mostly donated from zoos around the world
after an animal dies. They use DNA in 15-20% of their cases to link animals and
their human killers to a crime scene. In combination with ballistic evidence
and fingerprints they have become very accurate at tracking illegal hunters and
poachers. In one example a man returned home after an illegal hunt and washed
and dried his clothes at home. The lab was able to use animal hairs found in
the dryer’s lint holder to place him at the crime scene. They can also use
barely visible bloodstains on clothing to track hunters down years later, and
with absolute statistical certainty.
5. To study
endangered animal species:
Helpful
when trying to discover if an animal that has been found is really an
endangered species or only a mutant of another species.
6. Detecting
genetically modified organisms:
Used
especially for agricultural identification. Many of the crops currently grown
in the United States contain genes that were introduced in order to develop a
new type of crop. They can be detected in the crop using PCR. Approximately
50-75% of produce and processed foods in grocery stores in the United States
are genetically modified or contain genetically modified organisms.
7. Testing
for e. coli and other illnesses carried by food sources:
Done
with PCR, using primers specific to certain disease strains can test for
pathogens like e. coli or mad cow disease in food such as hamburger meat.
8. Paternity
and Maternity testing:
DNA
paternity testing is the most accurate way, with 99.9% certainty, to determine
paternity. It can be used for maternity testing also but that is quite uncommon
because mothers usually give birth at a hospital or with others that can
testify to her being the mother. (Example: maternity testing could be used on a
child that was born at home and then anonymously left at an orphanage or
hospital.) Such accurate testing is possible because a person inherits VNTR
patterns from their parents, and every persons DNA is unique except for that in
identical twins. VNTR patterns are extremely specific and can be accurately
compared. A child will share one band with the biological mother and one with
the biological father. They can be used for parent identification as well as
biological parenthood in adoption cases.
9. Personal
Identification:
This
application of DNA fingerprinting has been discussed but is not in use at the
current time. It would involve using the VNTR patterns of individuals as a type
of genetic bar code to identify them. This is relatively impractical because it
would be far too expensive and time consuming to analyze and store millions of
VNTR patterns to be used as personal identification references.

These alligator snapping turtles were recovered in 2002 in
Greenwood, Mississippi, in a sting operation that ended in the arrest of a
seller.
|
|
|
Confiscated animal products, including tusks,
tortoise shells, and stuffed birds, at the U.S. Fish and Wildlife National
Forensics Lab. |

![]()
|
Through the Grapevine Ringed
by his handiwork, Cornell researcher Bruce Reisch examines the leaves of a
Chardonnay grapevine for signs of fungal disease. Reisch and his colleagues
are working to create Merlot and Chardonnay grapes that are genetically
modified to resist diseases that now must be fought with fungicides. “Our
goal is to come as close as possible to eliminating the need for spraying,”
he says. |

![]()
|
Building a Better Tomato
A slide representing 20,000 tomato genes is projected onto Mark D’Ascenzo, a researcher at Boyce Thompson Institute for Plant Research in Ithaca, New York. Scientists there are trying to identify the genes that make certain tomatoes resistant to diseases. "We’ve isolated hundreds of genes that are interesting candidates," D’Ascenzo says, "but we’re still years away from understanding the whole picture." Once scientists do, the genes that are responsible for resistance can be synthesized and inserted into a new generation of tomato plants. |
http://www.sumanasinc.com/webcontent/anisamples/dynamicillustrations/paternitytesting.html
Case Studies
Murder/Rape Case: State of Florida vs. Jones and Reesh
Murder at Rodman Dam, 1988
In
July of 1987, teenagers Randall Scott Jones and Chris Reesh were target
shooting with a 30/30 hunting rifle at the Rodman Dam recreation area in
Florida. While they were shooting,
Jones’ pickup truck became stuck in a sandpit.
A fisherman suggested that the two ask for help from a nearby pickup
truck. The two approached the truck
where they found Kelly Lynn Perry and her fiancé, Matthew Brock, sleeping. Jones and Reesh debated on whether or not
they should wake the two to ask for help.
The
next morning, fishermen found the bodies of Perry and Brock in the woods next
to the recreation area. Police
investigation showed that each was shot in the head with a 30-caliber
bullet. Brock was shot twice in the
head, and Perry was shot once. It was
also found that Perry had been sexually assaulted. The pickup was reported stolen.
In
August, Jones was arrested in Mississippi driving Brock’s truck. Reesh was arrested the next day after Jones
had said the two were together that night in July. Both were indicted on counts of first-degree murder and sexual
battery.
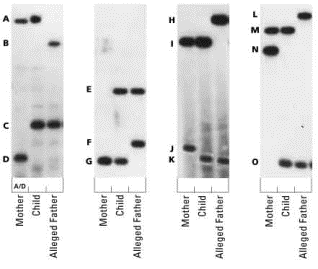
A
semen sample was taken from Perry’s body, and blood samples from Jones and
Reesh were taken. They were compared in
a laboratory that specializes in DNA fingerprint testing. The DNA fingerprint indicated that Jones had
raped Perry.
Using
DNA fingerprinting along with other evidence, events of the crime were put
together. Jones confessed to shooting
both Perry and Brock in the head at close range. The two teens then dragged the bodies into the woods. They towed Jones’ truck out of the sandpit
with Brock’s truck, and the two drove off with both trucks. Later, Jones returned to the bodies, took
them further into the woods, and raped Perry.
A
representative from the DNA fingerprinting laboratory testified that the chance
of another person having the same DNA fingerprint as Jones was one in
9,390,000,000.
The
jury only deliberated for 30 minutes.
The jury convicted Jones of two counts of murder, one count of burglary,
one count of shooting into an occupied vehicle, and one count of sexual
battery. The judge sentenced him to a
double death sentence. This made the
case the first in which DNA fingerprint evidence was used, and the death
sentence was given. Reesh was sentenced
to six years in prison and twenty years probation.
Human Paternity Case: Sarbah vs. Home Office
Ghana Immigration Case, 1985
The first practical test of DNA
fingerprinting involved Christina Sarbah and the Home Office in England. Christina wanted to prove that Andrew, who
had been living in Ghana with Christina’s estranged husband, was indeed her
son.
Immigration officials held Andrew at
Heathrow Airport, because they claimed that his passport was forged, or a
substitution had been made. Member of
Parliament Martin Stevens intervened and made it so the child could stay at his
family’s home in London with siblings David, Joyce, and Diana.
The Hammersmith Law Centre, who
provides legal aid to the underprivileged, put together large amounts of
evidence, including pictures and statement by family members. Various tests to determine genetic
characteristics showed that Christina and Andrew were almost certainly
related. What the tests did not prove
was whether or not Christina was the mother or merely an Aunt. This evidence was rejected at an immigration
hearing, but deportation was put off pending an appeal.
Centre
workers contacted Alec Jeffreys at Leicester University after they read in the
local newspaper about a scientific discovery that could prove maternity, and
they asked him to take on the case.
Jeffreys accepted the case because he believed it would be an ideal test
of the DNA fingerprint technology he had recently developed. He compared the DNA extracted form the blood
samples from Christina, Andrew, the three other children, and an unrelated
individual. The resulting DNA
fingerprint would verify whether or not Andrew is Christina’s son.
It
was questioned whether Christina was Andrew’s Aunt, a sister of his true
mother. Andrew’s fingerprint contained
about 25 bands that were inherited from his mother. The possibility that Christina is the true mother’s sister and
yet happens to share 25 bands with her is about one in 600,000.
The
case was complicated because of the lack of the father’s blood sample and the
doubts about Andrew’s paternity.
Jeffreys reconstructed the father’s fingerprint from bands present in
the three undisputed children, but absent in Christina. About half of Andrew’s bands match bands in
the father’s compilation, and the remaining bands were all present in Christina’s
fingerprint. The possibility of this
happening by chance is greater than one in a trillion.
Horse Paternity Case: Thoroughly Bred
Father or Son? 1989
Thoroughbred
horses are bred primarily for galloping speed, which is based on a number
inherited characteristics, including muscle mass, conformation, and
cardiopulmonary capacity. These animals
are carefully mated, and their ancestry can be traced back many
generations. Verification of an
animal’s parentage is key to its value.
This
case involves a leading stud horse, from one of the most prominent thoroughbred
lines, and his son. The father was
retired from studding when low sperm count indicated he was becoming
infertile. Mares without foal who had been
mated to the father were then remated to the son. However, standard blood tests could not confirm whether father or
son was the sire of the foals conceived during this changeover period.
The
autoradiogram proved that the son sired the foal. Although the foal shared a band with the father, all of the
foal’s bands are accounted for only by including two that originated with the
son.
As a test of identity, DNA fingerprinting can
positively determine thoroughbred parentage.
However, DNA fingerprinting of horses is far more difficult than it is
in humans. Due to centuries of meticulous
inbreeding, all thoroughbreds are derived from a closed pool of genetic
material. Even thoroughbreds not
directly related share a common genetic heritage, and consequently, may also
share a large number of DNA fingerprint bands.
Bibliography
Russell, P.J.
(2002). iGenetics. San Francisco: Benjamin Cummings.
Sheindlin, Judge
Gerald. Genetic Fingerprinting: The Law and Science of DNA. Bethel, CT:
Rutledge Books, Inc., 1996.
On the Human Genome Project. http://homepage.smc.edu/hgp/history.htm#timeline
Basics of DNA Fingerprinting. http://www.biology.washington.edu/fingerprint/dnaintro.html
DNA I.D. http://whyfiles.org/014forensic/genetic_foren2.html
MSN Encarta. http://encarta.msn.com/encyclopedia_761579857/DNA_Fingerprinting.html#endads
Kary Mullis’ Biography. http://www.karymullis.com/bio.html
Dolan DNA Learning Center: DNA
Fingerprinting. http://www.dnalc.org/resources/aboutdnafingerprinting.html
DNA Fingerprinting in Human Health and
Society. http://www.accessexcellence.org/AB/BA/DNA_Fingerprinting_Basics.html
How DNA
Evidence Works. http://science.howstuffworks.com/dna-evidence3.htm
http://www.biotech.iastate.edu/biotech_info_series/bio7.html#anchor15180002
http://edition.cnn.com/2004/TECH/science/05/22/high.tech.poaching.ap/
http://www.ornl.gov/sci/techresources/Human_Genome/elsi/gmfood.shtml
http://www.geneseek.com/link.sp?page=ident-parent-cattle
http://www.biology.washington.edu/fingerprints/apps.html
http://www.nationalgeographic.com
http://www.dnalc.org/shockwave/dnadetective.html
